Biological systems are often described by pairwise interactions between components, such as protein interactions within a cell or species interactions in an ecosystem. However, unlike physical systems where the interactions are determined by fundamental forces with universal coupling constants, the interaction strengths between biological components can be modified by third parties, such as the allosteric regulation of protein interactions. That is because biological components have complex internal structures, and the effective strengths of their interactions can change if the internal structures are altered.
-
trait-mediated effects in ecology
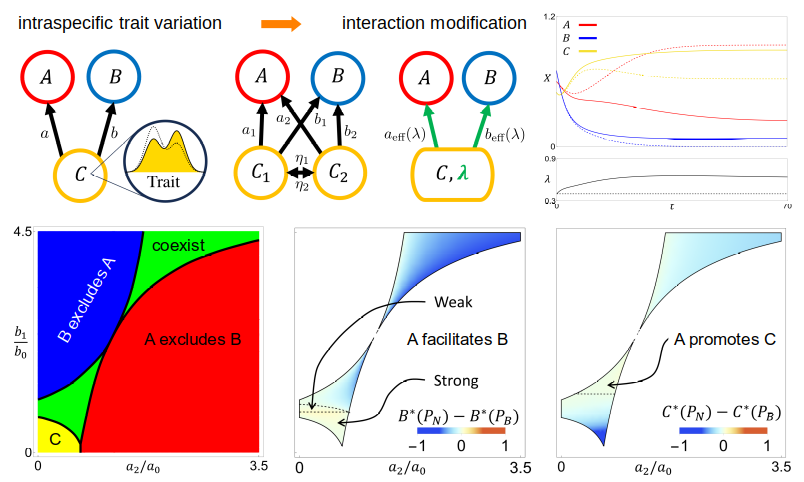
Traditional models of ecosystems assume a constant interaction strength between a pair of species. However, individuals of the same species vary in traits that can affect how they interact with the environment or other species. The distribution of traits may shift under the influence of other species, leading to the modification of interaction strengths. As an example, we analyzed an ecosystem with two predators and a shared prey [ref], which would result in the competitive exclusion of one predator if the interaction strengths are fixed. We showed that when there is trait variation within the prey that can shift dynamically, the ecosystem can stabilize at a new equilibrium where all three species coexist. We also uncovered trends that are unexpected in models with constant interaction strengths, such as the facilitation of one predator by the other (i.e., the abundance of one predator is increased when the other is present) and the promotion of the prey by one predator (i.e., the abundance of the prey is increased when the predator is present). These “abnormal” trends have broad consequences on many types of ecosystems and larger networks.
-
robust retrieval of dynamic sequences
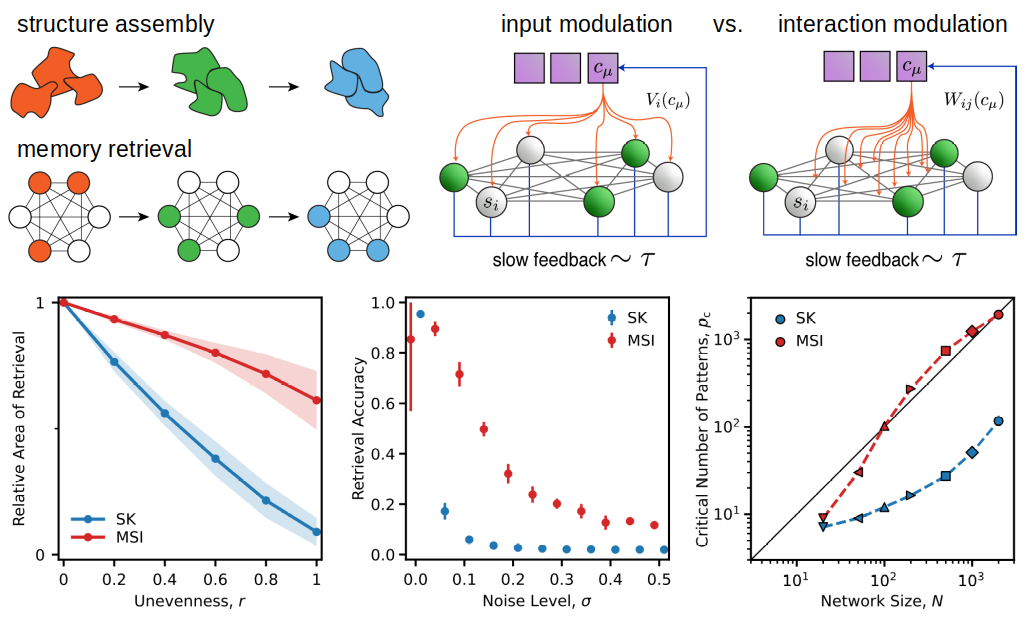
Many biological systems are able to dynamically rearrange their components through a sequence of configurations in order to perform their functions, such as neurons altering their synaptic strengths to generate a sequence of activity patterns, or protein assemblies rearranging their composition to go through several stages of processing substrates. Such dynamic processes have been studied using network models that sequentially retrieve a set of stored patterns by controlling inputs to the network components (“input modulation”). In contrast, we introduced a new class of models that can retrieve dynamic sequences by modifying the interactions among the components (“interaction modulation”) [ref]. We showed that interaction modulation models can robustly retrieve patterns with different activity levels and have a much larger dynamic storage capacity. Such interaction modulation may be a new paradigm for modeling complex collective dynamics.